Home Research Centers Plant Cytometry Laboratory

Plant Cytometry Laboratory

News article in 1988 (Nebraska Alumnus magazine)
David Galbraith’s laboratory pioneered the application of the techniques of flow cytometry and cell sorting to that most important kingdom of life, the plant kingdom. Starting in 1980, with one of the first Coulter EPICS V flow sorters to be placed in an academic laboratory, Dr. Galbraith and his co-workers started to address the confounding question as to how to render three-dimensional, multicellular plant tissues and organs suitable for analysis using instruments compatible only with single cell suspensions. One successful approach involved the analysis and sorting of protoplasts (plant cells from which the cell walls had been enzymatically removed, hence separating the cells into cell suspensions). However, since plant cells are typically larger, and sometimes much larger than the blood cells around which flow instrumentation was initially designed, this raised the immediate problem of how to implement flow analysis and sorting using flow tips that were correspondingly larger in diameters than the 50-70 µm size typically employed with animal cells.

News article in 1988 (Nebraska Alumnus magazine)
Successful sorting using flow tips having diameters of up to 200 µm required lowering the system pressure as well as the droplet generation drive-frequency, modifying the point of view of droplet break-off, and defining suitably indestructible standard particles of similar sizes for sort optimization. This developmental work aimed at providing the ability to detect and sort heterokaryons based on induced fusion of protoplasts, and with the accompanying development of methods for fluorescent labeling of the input protoplasts, it became possible to sort and regenerate these heterokaryons to produce somatic hybrid plants. These methods were the basis of an issued U.S. patent, and have been employed for agricultural improvement of particular crops, notably Brassica napus (canola or rapeseed), as well as for basic studies of the influence of cell size on cellular metabolism, physiology, and cell biology. The flow methods also are applicable for the sorting of additional examples of large biological cells, and are increasingly being employed for mammalian systems. Correspondingly, the instrument manufacturers now routinely include modified hardware (flow tips, etc.) and operational parameters for large particle sorting as sales features.
Picture of David Galbraith circa 2000 with MoFlo
A second successful approach addressed means for the analysis of nuclear DNA content and cell cycle status within plant tissues using flow cytometry. We were able to show that protoplasts could be prepared, fixed, stained with DNA-specific fluorochromes, and analyzed to give recognizable DNA histograms. We also found G1 peak CVs were greatly improved by adapting detergent-based treatments for simultaneous protoplast lysis and nuclear DNA staining. However, the whole field really took off when we defined a simple chopping procedure to release nuclei from whole plant tissues and organs, followed by DNA-specific fluorochrome staining and flow analysis. This simple and rapid procedure is now routinely employed world-wide for measuring nuclear DNA contents of different plant species. It is also used for ploidy determination, with extensive applications in basic and applied plant sciences, botany, and agriculture. It has largely replaced other methods, including chromosome counting, and Feulgen staining, based on issues of accuracy, convenience, and cost.
More recently, we have been developing methods of flow cytometry and sorting to address the question of extracting molecular information contained within the heterogeneous cell types found in plant tissues and organs. This has involved three steps, pursued in parallel: devising fluorescent markers for specific cell types, providing the means for global extraction and analysis of molecular information, and developing the procedures to selectively enrich for the contribution of one particular cell type. Most successful in terms of marking specific cell types has been the Fluorescent Protein (FPs) of which the Green-Fluorescent Protein (GFP) is the prototype. We were amongst the first to demonstrate expression of GFP in plant cells, visualizing this using confocal microscopy. We also pioneered use of flow cytometry and cell sorting for the analysis and selective purification of GFP-expressing protoplasts. In further work, we developed methods and DNA constructions that target GFP to the nucleus of transgenic plants. For global information extraction, we employed DNA microarrays, and were one of the first to describe the production and use of microarrays for global gene expression profiling in higher plants.
Microarray Printer
We expanded our microarray printing activities to a number of different species from both the plant and animal kingdoms, and to date have distributed world-wide approximately 40,000 microarrays (containing on average 30,000 array elements). Most of these comprise synthetic 70-mer single stranded DNA array elements. For selective enrichment we have adopted two strategies. One is simply to flow sort GFP labeled protoplasts, utilizing cell type-specific promoters to drive expression of GFP only within particular cells. This work, in collaboration with the laboratory of Philip Benfey at Duke University, led to the description of the arabidopsis root in terms of a gene expression map.
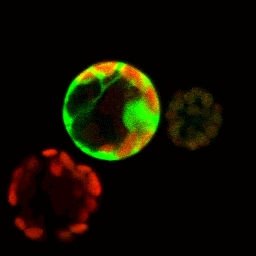
GFP-expressing protoplast used for flow sorting.
The second strategy involves targeting GFP to the nucleus, preparing cellular homogenates, and flow sorting the green-fluorescent nuclei. Nuclear transcripts are then globally characterized using microarrays. We identified protein partners suitable to confer nuclear retention of GFP in cell free homogenates. Placing these chimeric GFP fusion proteins under the transcriptional control of the promoters from genes expressed in a cell type-specific manner then highlights the nuclei only of these cell types. We have established that nuclear transcript levels faithfully recapitulate total cellular transcript levels, and that the sorting approach allows precise definition of global gene expression within minority cell types of the arabidopsis root. These techniques are applicable, evidently, to any eukaryotic organism, and we are extending their use to mammalian tissues and organs.
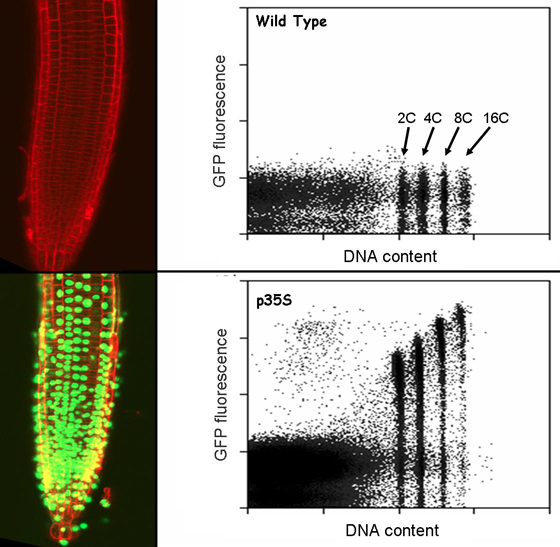
GFP-expressing plant root, and control, with flow histograms of nuclei produced from these samples.
Over the years, I have been active in a number of international training activities in flow and image cytometry, including courses and workshops offered in Germany, Poland, Italy, the Czech Republic, England, and Sweden. In the United State, I participate in teaching the annual flow course at Los Alamos and Bowdoin College. If one includes microarrays as an indirect form of image cytometry, we have offered at Tucson eight practical workshops over the last five years in the use of this technology, and have trained around 300 participants, drawn world-wide.

Microarray Workshop 2004
Current research activities include continuing our studies in cell type-specific gene expression, developing microarray platforms for high throughput analysis of proteins, developing low cost genotyping microarrays for plant species, and exploring the use of chemical genomics for selectively perturbing gene expression within plants and animals. Our work is highly collaborative, and has involved scientists drawn from many different universities and research institutions within the U.S. and across the globe. International collaborations currently involve the Czech Republic, China, and various CGIAR institutes (the Philippines, Colombia). We welcome visitors to the laboratory for collaborative work involving flow analysis and sorting, microarrays and image cytometry. Current major equipment includes a Cytomation MoFlo, a BioRad MP1024 confocal/multi-photon microscope, Omnigrid OG100 and OG300 and Genetix ArrayMax microarray printers, and Molecular Devices Genepix 4000B and 4200AL microarray scanners.

Confocal in the lab.

Microarray Printer (OG300)

David Galbraith skydiving circa 2004
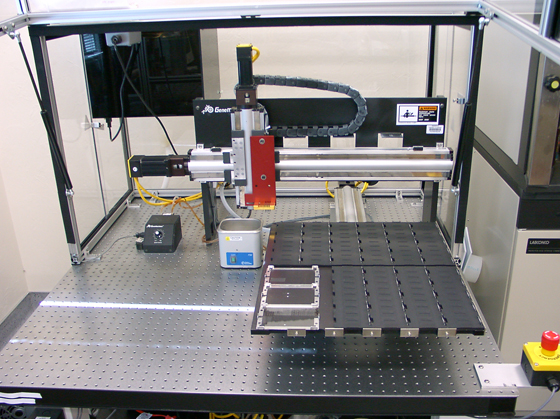
Microarray printer (OG100) in the lab
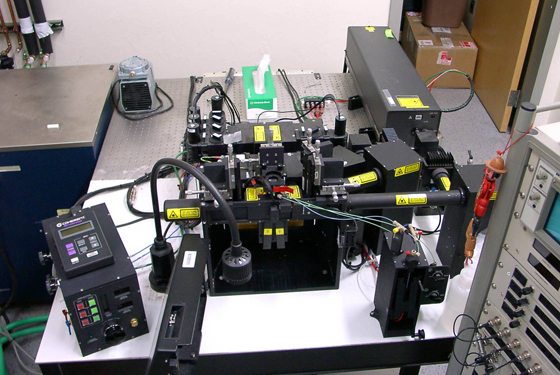
MoFlo in the lab
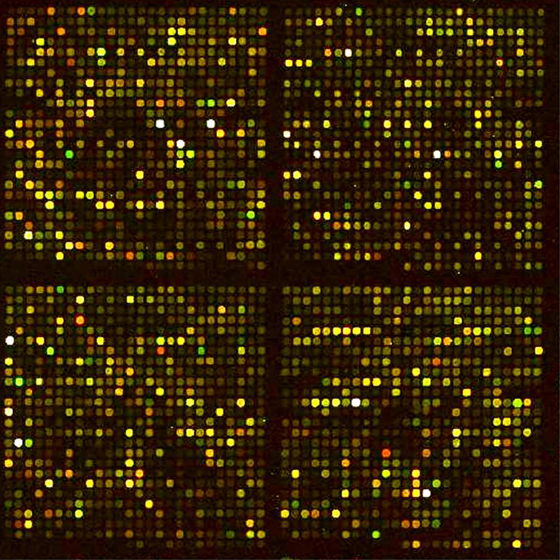
Image of a plant microarray following hybridization

Image of protein microarray showing GFP elements
