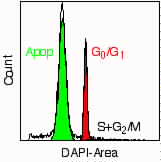
>I would like to get some information from anyone who is successfully using >the Modfit DNA Analysis program for analysis of DNA content, cell cycle, >and especially apoptotic populations. We are having quite a bit of >difficulty getting useful information from this program regarding apoptotic >populations. We are using mostly PI staining of nuclei for analysis. We >are also using cell lines, not clinical samples, so there is limited >involvement of an aneuploid population. The main trouble is setting the >markers so the data makes sense. We either see an apoptotic population >under the G1 and S phases of the histogram or the apoptotic population is >labeled as debris. We have no consistency in our data with regard to >apoptosis. The manual is not helpful. If anyone out there can help, it >sure would be appreciated. Best regards, Susan Zunino > >Dr. Susan J. Zunino >Lehrstuhl fur Genetik >University of Erlangen >Staudtstr. 5 >91058 Erlangen >Germany >Tele. 49 9131 8528784 >FAX 49 9131 8528526 >e-mail: szunino@biologie.uni-erlangen.de Dr. Zunino, Most likely the problem is not with ModFit, but with the data you wish to analyze. We were fortunate to get good results using either DAPI or PI staining for DNA content and observing a hypo-diploid peak with murine lymphocytes, where the fraction of cycling cells (S+G2/M) was low and the fraction of apoptotic cells was significant (after a short culture) [see attached JPEG, viewable with any web browser]*. I would not recommend this technique for cultured cell lines, since the fraction of cycling cells is significant and "apoptosing" G2/M phase cells would still have the same DNA content as healthy G0/G1 or even S phase cells. A better method would be TdT/dUTP identification of DNA "strand breaks" or "nicks" (sometimes called the "tunnel" assay). This is usually coupled with PI staining for DNA content measurements. Several companies offer kits, like Pharmingen's APO-DIRECT, which includes positive and negative control cells. Perhaps there are other methods that Dr. Zbigniew Darzynkiewicz could recommend. ModFit behaves as you describe above when it cannot discern a "floating" gaussian peak from background noise. If you can't visually see the peak, ModFit will not "see" it either. You can introduce some artificial dependencies between the apoptotic "peak" and the G0/G1 peak to improve consistency, but you will only generate worthless artificial results. Good luck, Eric *At 5350 byes, the JPEG should only take 3.7 seconds to download at 14400 bps.
 /\/\/\_ Eric Van Buren, aa9080@wayne.edu
\ \ \ Karmanos Cancer Institute and Immunology & Microbiology
\_^_/ Wayne State University, Detroit, Michigan, USA
/\/\/\_ Eric Van Buren, aa9080@wayne.edu
\ \ \ Karmanos Cancer Institute and Immunology & Microbiology
\_^_/ Wayne State University, Detroit, Michigan, USA
This archive was generated by hypermail 2b29 : Wed Apr 03 2002 - 11:53:47 EST