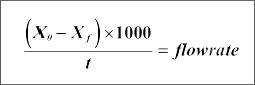Enumeration of aquatic viruses by nucleic acid-specific staining
in combination with flow cytometry
Dr. Corina Brussaard
Dept. Biological Oceanography
Royal Netherlands Institute for Sea Research
PO Box 59, NL-1790 AB Den Burg, Texel, The Netherlands
Corresponding author: corina.brussaard@nioz.nl
Solutions
* Fluorescent dye working solution SYBR Green I (Molecular Probes Inc.): 200 times diluted commercial stock
* Sample fixed with 0.5 % glutaraldehyde (EM grade, 25% solution). See Support Protocol 1
* TE-buffer 10:1 ; pH = 8.0 and 0.2 µm pore-size filtered
* Beads for internal standard: 1 µm beads (Molecular Probes, FluoSpheres carboxylate modified microspheres, 1.0 µm, yellow-green fluorescent). See Support Protocol 2
* MilliQ (freshly prepared)
Materials
* Flow cytometer (FCM) with 488 nm Argon laser : Becton Dickinson FacsCalibur for example
* FCM tubes
* Adjustable pipettes plus tips
* Tube racks
* Sterile 0.2 µm pore-size filters (e.g. FP30/0.2 µm Schleicher & Schnell)
* Tissues
* Waterbath 80oC and 37oC
Prestart
* Turn on a 80oC and 37oC waterbath
* Remove SYBR Green I working stock solution from freezer and let it thaw in the dark at room temperature
* Fill sheath fluid container from FCM with MilliQ. Switch on FCM and make sure the machine is very clean! Set trigger on green fluorescence. Make sure that the voltage stays below the level at which electronic noise appears!
* Determine the flow rate (See Support Protocol 3)
Procedure
* Take samples (e.g. 8 per series) out of the - 80°C freezer and let them thaw in the 37°C waterbath. The sample must stay cool!
* Label tubes
* Add 5 µl of beads working stock solution to the tubes (See Support Protocol 3)
* Dilute your samples in TE. Add one tube as blank with TE and sterile 0.2 µm pore-size filtered seawater
* Dim light and add SYBR Green I dye to a final concentration of 0.5 10-4 of the commercial stock
* Incubate 10 minutes at 80oC in the dark
* Allow the sample to cool in the dark before analysis (about 5 min)
* Run your sample at appropriate flow rate. The number of events/sec must be between 100 and 1000. If not, use a different dilution or another flow rate. Blank should not have very low event rate!
* When ready, clean FCM and switch off FCM, as well as waterbaths. Put dye back in freezer (can be reused) and clean up
Results
Typically, 3 populations of viruses can be detected in natural samples;
differing in their green fluorescent signal (Figure 1).
Phytoplankton viruses may (not necessarily though) have flow cytometric
signatures that allow them to be discriminated from the remaining bulk
of viruses (Figure 1).
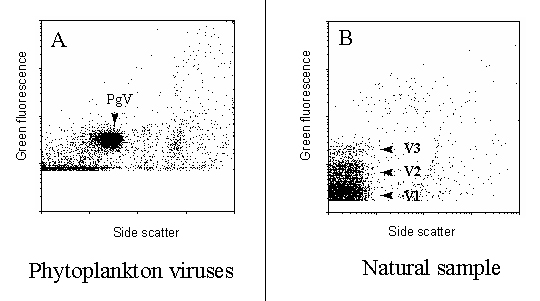
Figure 1 : Green fluorescence vs Side Scatter cytograms of phytoplankton
viruses and viruses from a natural sample.
To calculate the abundance of viruses in a sample, select for viruses and distract the viral count of the blank from the total viral count of the true sample. Correct for running time, flow rate and dilution to obtain final abundance of viruses per ml.
Support protocol 1: preservation and storage of samples
* Add glutaraldehyde to freshly obtained water samples at final concentrations of 0.5 %. Work in hood
* Fix for 15-30 minutes at 4oC
* Freeze the samples in liquid nitrogen (N2)
* Store at -80oC
* Before use: thaw samples quickly at 37oC. Measure as soon as possible, because the viral abundance goes down after thawing
* Save remaining sample cool (4oC) for max. a few hours (in case sample analysis went wrong, you can redo it)
Support protocol 2: internal standard beads preparation
Materials
* Sterile MilliQ water
* 1 µm beads (Molecular Probes, FluoSpheres carboxylate modified microspheres, 1.0µm, yellow-green fluorescent)
* Sterile tube with lid
Procedure* Mix beads very well before use (eventually, sonicate briefly).
* Prepare primary stock by adding 1-2 drops of the commercial stock in 10 ml sterile MilliQ in tube. Store at 4 C (check for quality on FCM prior to addition to samples)
* Prepare working stock by adding 10 µl primary stock to 2.5 ml sterile MilliQ in sterile tube with lid. Can be used during full day. Prepare new for next day
Support protocol 3: calibration of flow rate for Becton
Dickinson flow cytometer
* Have MilliQ as sheath fluid
* Select a flow rate
* Fill a tube with TE
* Weigh the sample (Xo)
* Remove the outer sleeve of the injection system carefully
* Wait until a droplet falls. Before the next one forms, put on the sample tube and close the sample arm in the running position. Simultaneously, start the chronometer
* Run the sample for at least 15 minutes
* Remove the tube and simultaneously stop the chronometer
* Weigh the remaining volume
* Calculate the flow rate using the following formula:
With :
Flow rate = µl/min
Xo = initial weight
Xf = final weight
t = time (min)
Acknowledgements
This protocol was reproduced from Brussaard et al. (2004) with kind
permission of the American Society for Microbiology.
Reference
Corina P. D. Brussaard.Optimization of Procedures for Counting Viruses
by Flow Cytometry. Applied and Environmental Microbiology. Vol 70 (3),
2004: 1506–1513