Association of TSA-FISH ( Tyramid Signal Amplification of Fluorescent In Situ Hybridization) and flow cytometry to quantify picoeukaryotes in the natural environment
Isabelle C. Biegala
Institut de Recherche pour le Développement, Centre Océanologique
de Marseille, rue de la Batterie des Lion, 13007 Marseille, France
Corresponding author: isabelle.biegala@free.fr
Introduction.
Picoeukaryotes (cells < 3 µm) contribute significantly to marine
plankton biomass and productivity, and recently molecular studies have
brought to light their wide diversity (Li 1994, Moon Van Der Staay et
al. 2001). Among the methods that have been used so far to quantify aquatic
microorganisms, fluorescent in situ hybridization of oligonucleotide probes
combined with flow cytometry offer both the advantages of high resolution
for taxonomic identification and automated cell counting (Amann et al.
1990, Wallner et al. 1993). However, cell losses, cell clumps and low
level of signal to background ratio have often been mentioned as major
problems for routine application of this combination of techniques (Lebaron
et al. 1997, Schönuber et al. 1997).
Method.
A new method is presented that combine TSA-FISH of probes targeting
16S rRNA and flow cytometry. The main points are the following:
1) The TSA enhancement of the fluorescent signal for FISH was essential for the detection of picoeukaryotes, as these small cells have a natural autofluorescent background that prevent them to be detected by classical FISH techniques. In addition, in the natural environment both exponentially and stationary growing cells can be encountered which have variable amount of rRNA and makes TSA fluorescent enhancement system necessary.
2) The use of surfactant (such as Pluronic F68 and dimethyldichlorosilane) and ultrasonication proved to be essential for avoiding cell loss and cell aggregation respectively, which prevent from precise cell quantification.
3) Picoplanktonic cells are collected with the help of surfactant by centrifugation from as little as few tenth of milliliters of 3 µm pre-filtered sea water without significant cell loss.
Results.
The routine application of the technique was tested along a coastal
transect off Brittany (France; Figure 1), where the different groups of
picoeukaryotes were investigated using already published probes and a
newly designed probe that targets the order Mamiellales (Prasinophyceae,
Chlorophyta; Figure 2).
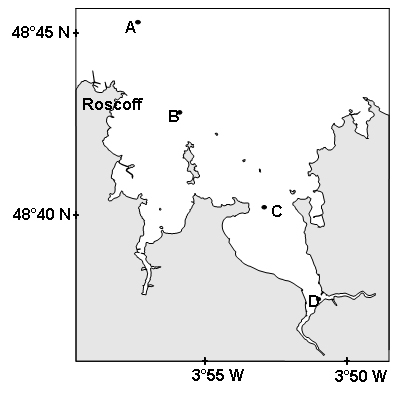
Figure 1. Localization of the different stations along the transect realized on July 17, 2002, in the Bay of Morlaix on the coast of Brittany (France).
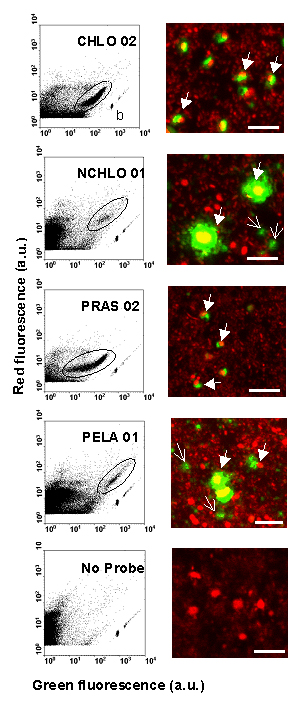
Figure 2. Cytograms and confocal microscope images on natural picoeukaryotes community from station D (Fig. 1) hybridized with divisions (CHLO 02 for Chlorophyta and NCHLO 01 for non Chlorophyta), class (PELA 01 for Pelagophyceae, Chlorophyta) or order (PRAS 02 for Mamiellales, Chlorophyta) specific probes and without probe (control). The cell hybridized probes were colored (TSA-FISH technique) with fluorescein (green fluorescence) and cells DNA were stained with propidium iodide (red fluorescence). Positively hybridized populations are encircled. b = 0.95 µm fluorescent beads. Closed arrows = probe labeled cells. Open arrows = unidentified particles. Bars = 5 µm. a. u. = arbitrary units.
Among the picoeukaryotes, the Mamiellale order of the Chlorophyta division out-numbered by one order of magnitude both the cyanobacteria and the non-Chlorophyta divisions, which were mainly represented by the class of Pelagophyceae (Figure 3).
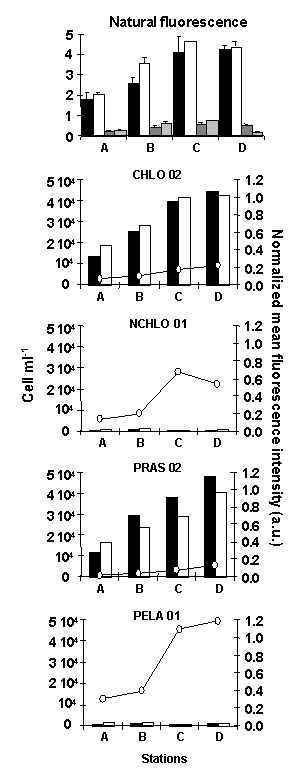
Figure 3. Cell counts obtained on natural picoeukaryote communities sampled at the different stations A, B, C and D along a transect in the Bay of Morlaix (Brittany, France, Fig. 1). The top panel shows a comparison of cell counts, based on the natural fluorescence of picoeukaryotes (black and white bars) and cyanobacteria (grey bars), between two protocols: concentrations of fixed cells by centrifugation prior to hybridization (black and dark grey bars) vs. cell fixation without centrifugation (white and light grey bars). The bottom four panels show a comparison of cell counts and mean fluorescence intensity of hybridized cells with different probes (see legend Fig 2). Cells were counted either by flow cytometry (black bars) or by fluorescence microscopy (white bars). a.u. = arbitrary units.
Picoeukaryote concentrations were increasing from open towards more estuarine waters (Figure 3), following probably changes in water temperature and stability.
For more information on this work see Biegala, I. C., F. Not, D. Vaulot and N. Simon. 2003. Quantitative assessment of picoeukaryotes in the natural environment by using taxon-specific oligonucleotide probes in association with tyramide signal amplification-fluorescence in situ hybridization and flow cytometry. Appl. Environ. Microbiol. 69:5519-5529
Acknowledgements
Figures were reproduced from Biegala et al. (2003) with kind permission
of the American Society for Microbiology.
Cited references
Amann, R. I., L. Krumholz, and D. A. Sthal. 1990. Fluorescent-oligonucleotide
probing of whole cells for determinative, phylogenetic, and environmental
studies in microbiology. J. Bact. 172:762-770.
Biegala, I. C., F. Not, D. Vaulot and N. Simon. 2003. Quantitative assessment
of picoeukaryotes in the natural environment by using taxon-specific oligonucleotide
probes in association with tyramide signal amplification-fluorescence
in situ hybridization and flow cytometry. Appl. Environ. Microbiol. 69:5519-5529
Lebaron, P., P. Catala, C. Fajon, F. Joux, J. Baudart, and L. Bernard.
1997. A new sensitive , whole-cell hybridization technique for detection
of bacteria involving a biotinylated oligonucleotide probe targeting rRNA
and tyramide signal amplification. Appl. Environ. Microbiol. 63:3274-3278.
Li, W. K. W. 1994. Primary production of prochlorophytes, cyanobacteria,
and eucaryotic ultraphytoplankton: measurements from flow cytometric sorting.
Limnol. Oceanogr. 39:169-175.
Moon-van der Staay, S. Y., R. De Watchter, and D. Vaulot. 2001. Oceanic
18S rDNA sequences from picoplankton reveal unsuspected eukaryotic diversity.
Nature 409:607-610.
Schönuber, W., B. Fuchs, S. Juretschko, and R. Amann. 1997. Improved
sensitivity of whole-cell hybridization by the combination of horseradish
peroxidase-labeled oligonucleotides and tyramide signal amplification.
Appl. Environ. Microbiol. 63:3268-3273.
Wallner, G., R. Erhart, and R. Amann. 1995. Flow cytometric analysis
of activated sludge with rRNA-targeted probes. Appl. Environ. Microbiol.
61:1859-1866.