Cytomics
Current info on Cytomics... www.Cytomics.info
J. Paul Robinson, Ph.D.
|
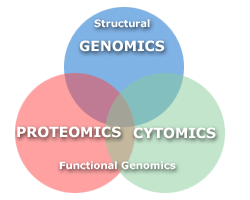
What is Cytomics? Functional relationships between the cell (Cytome) and the metabolic pathways (Proteomics-proteome) resulting from genetic control mechanisms (Genomics-genome) - some relate Cytomics to what is being termed functional genomics. |
|
Who does Cytomics?
Cytomics will require the specialized technologies of cellular analysis tools such as flow cytometry, image analysis, confocal and other single cell analytical techniques
Where do I find information about Cytomics?
Go to the Cytomics web server at www.cytomics.info
First Proposed: (related to plant studies)
proposed by Davies E, Stankovic B, Azama K, Shibata K, Abe S. Novel components of the plant cytoskeleton: A beginning to plant "cytomics" Plant Science. Invited Review, Plant Science (160)2 (2001) pp. 185-196.
http://web-mcb.agr.ehime-u.ac.jp/english/cellbiol/default.htm
What do others say about Cytomics?
http://www.ipd.anl.gov/biotech/programs/func-genomics/
Cytomics (an ad hoc term, used for convenience) provides much of the scientific challenge anticipated in genomics research; it will occupy biological researchers for many years. At one level, in its goals and scope, functional genomics is not so different from the traditional field of cell biology. The difference lies in the fact that this research is now transformed by the more global perspective provided by the genomic "satellite system." http://www.geocities.com/florecitafolmer/paper08.htm
Proteomics consists in the study of the proteome. The term proteome was introduced for the first time by Marc Willens in 1994 and it can be defined as the complete set of proteins encoded by the genome of a given organism (Neumann, 2001). Proteomics is very closely related to functional genomics, and it is even considered by some authors, including Stephens (2001), as a subdivision of functional genomics, next to "cytomics", which is defined as the research area that encompasses the dynamics of the living cell.
http://www.biochem.mpg.de/valet/cytomics.html
cytomics: Multiparameter cytometric analysis of the cellular heterogeneity of cytomes, ... access a maximum of information on the apparent molecular cell phenotypes, resulting from cell genotypes and exposure. Molecular cell phenotypes in the naturally existing cellular and cell population heterogeneity of disease affected body cytomes contain the information on the future development (prediction) as well as on the present status (diagnosis) of a disease. [G. K. Valet, Predictive Medicine by Cytomics" Max- Planck- Institut für Biochemie, Martinsreid, Germany, 2001]
http://www.isac-net.org/enews/Summer01/world.htm
Measurements at the level of the cytome can be used to inform mathematical descriptions of the changes in complex biological systems. The cytome concept will start to drive the demand for existing and the quest for new single-cell analysis technologies. We will increasingly realize our ability to evolve cytome behavior into higher-order responses including those at tissue, organism and population levels. This offers the possibility of informed prediction from the culture dish to the clinic. ISAC [International Society for Analytical Cytology E-News, Summer 2001]
The bulk of our knowledge concerning the plant cytoskeleton has come primarily from the use of techniques and probes derived from animal research. However, in comparison with animal tissues, relatively few plant cytoskeleton proteins have been identified. We presume this is not because the plant cytoskeleton is really made up of such few proteins, but rather that only rarely have attempts been made to identify plant-specific cytoskeleton proteins, using plant-specific methods. Here we outline methods that we have developed both for the isolation and identification of novel cytoskeleton proteins as well as for the visualization of novel filamentous structures in plant cells, and we describe several novel cytoskeleton proteins and two novel cytoskeleton structures, 'nanofilaments' and 'nanotubules'. We postulate that use of such approaches will lead to a rapid expansion of our knowledge of the plant cytoskeleton. [E. Davies et. al. "Novel components of the plant cytoskeleton: a beginning to plant 'cytomics'" Plant Science 160(2): 185- 196, Jan. 5, 2001]