A. Salas1, C. Secades1, F. Sansonetty2,
M. Alonso-Guervós1, A. Sampedro1.
1Image Processing and Cytometry Service, University of
Oviedo, Spain.
2IPATIMUP, Instituto de Patologia e Imunologia Molecular
da Universidade do Porto, Portugal.
The reproducibility of the results of the DNA analysis by flow cytometry is conditioned by very diverse factors. Given the wide variability of methodology followed by the different cytometry laboratories, the exchange of data files allows evaluation of the differences that are due to individual interpretations of cytometric data and eliminates the variability in preparation and technical considerations of the analysis1. In 1984, Murphy and Chused proposed the first flow cytometry data file format FCS 1.0 that provided a uniform file format allowing data acquired on one computer to be correctly read and interpreted on other computers running a variety of operating systems. That standard, modified in 1990 and adopted by the International Society of Analytical Cytology, permits the establishment of interlaboratory data exchange to solve cases of difficult interpretation.
In the past, the usual way to exchange data files was through postal sending of diskettes previously agreed by the participants with consequential delay in the obtaining of results. Since the Internet network have been made accessible, the possibility of using it with the purpose of establishing quality assurance in DNA ploidy analysis have arose2. In the Work Group of Solid Tumors of the Iberian Society of Cytometry, a pilot trial was carried out for the distant study of DNA histograms between cytometry services from Oviedo in Spain, and Porto in Portugal.
1. Preparation of the pages
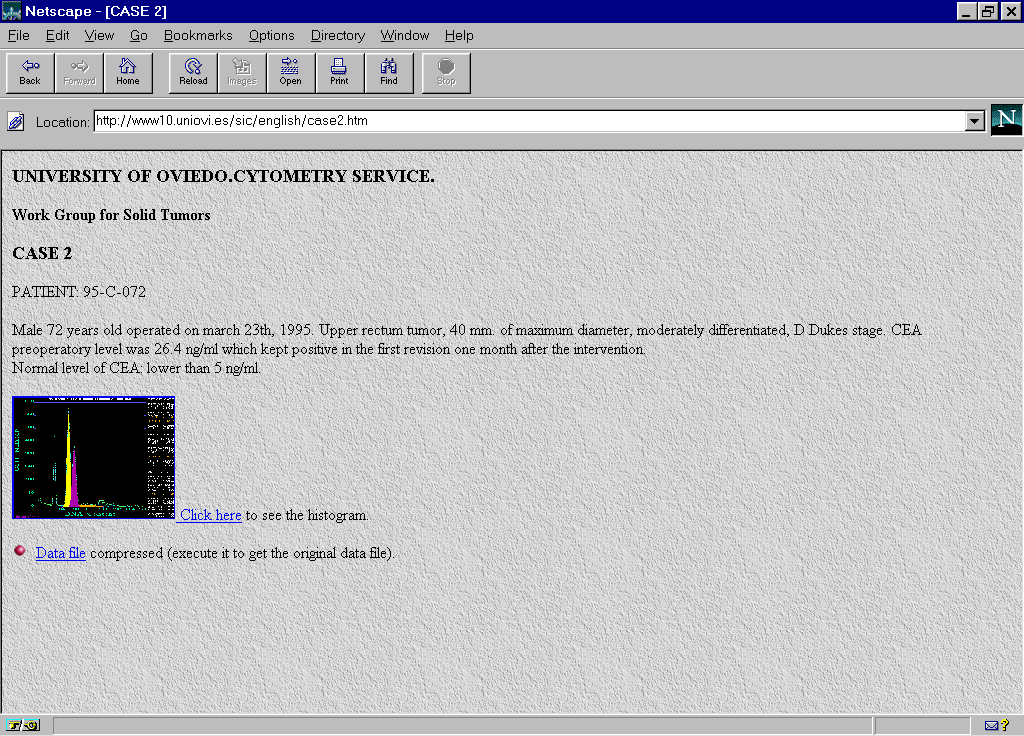
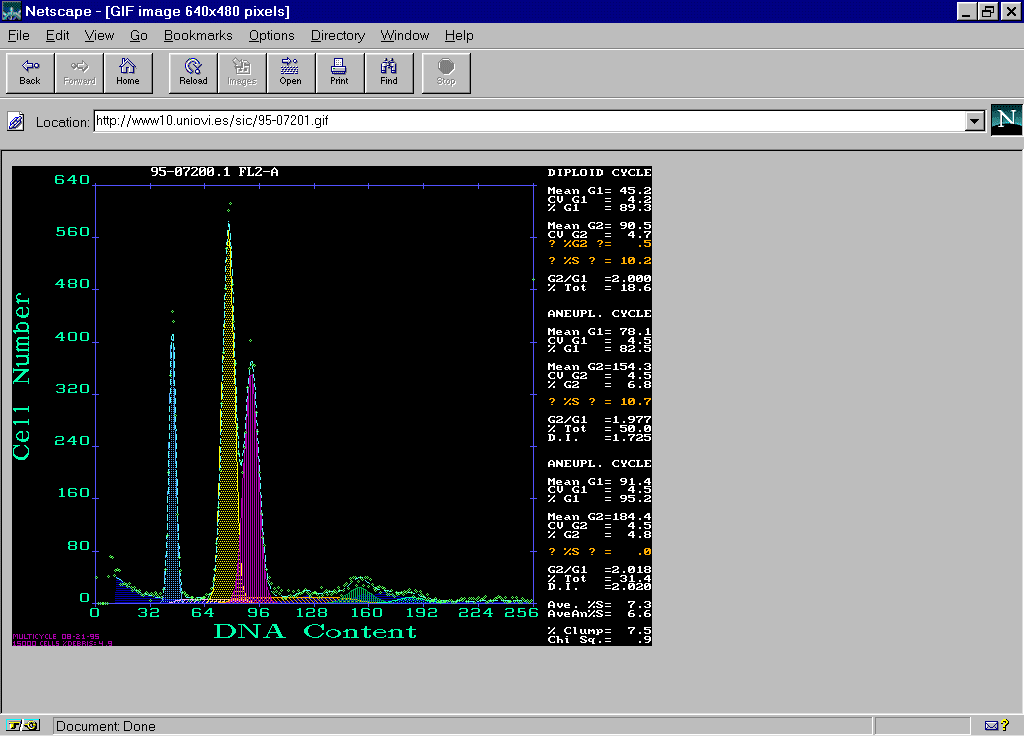
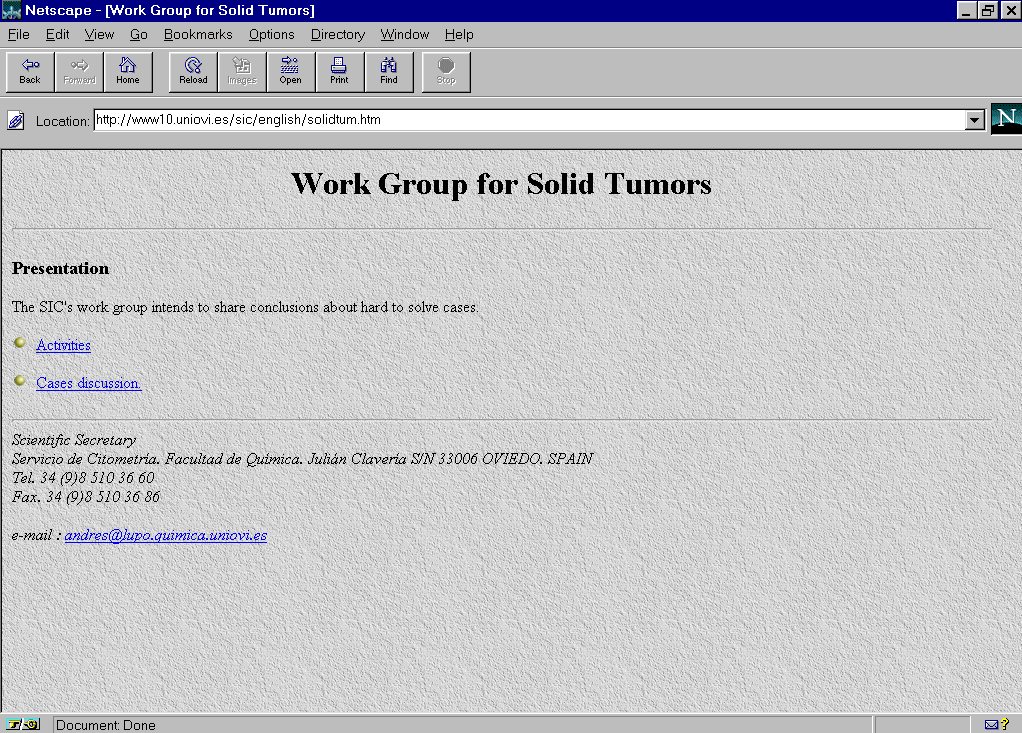
In the WWW server of the Image Processing and Cytometry Service in the University of Oviedo were edited pages for the Work Group of Solid Tumors (http://www10.uniovi.es/sic/english/solidtum.htm) (Fig. 1). Information corresponding to 10 cases of colon carcinoma of unclear interpretation were included.. For each case the technique followed in the preparation of samples, the conditions of analysis as well as clinical data were specified (Fig.2). Each case was accompanied by a histogram analyzed in the reference center by means of the Multicycle software from Phoenix Flow Systems with the numeric results and the cytometric data file (Fig.3, Fig 4).
A link to the data by a self-extracting data file (.EXE) was prepared for each case. The files were compress by the software program RAR archiver which produce a compressed file to execute. When it is executed the original file is obtained.
2. Access to the files of cytometric data
The client or recipient, clicks on the icon and file automatically the information on the disk drive. Once the file is executed the data is ready for analyzing by the specific DNA software and for obtaining the new results from each individual criteria.
3. Sending the results
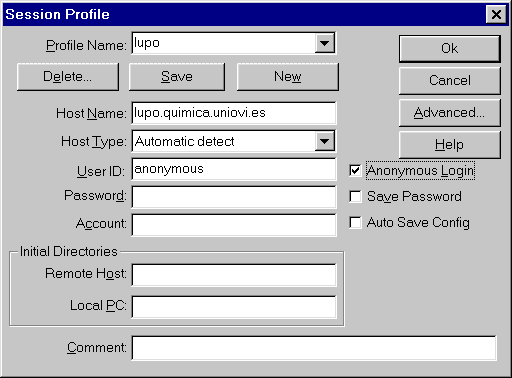
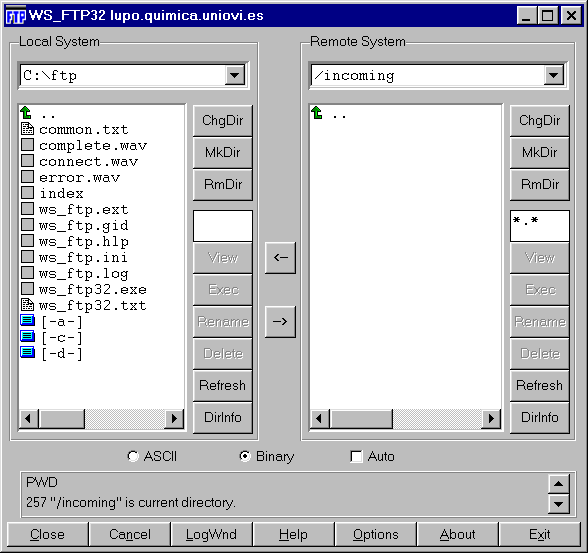
In this first experience, as soon as the participant analyzes the cases, the results file was sent to the reference center in Oviedo. This transmission of files was carried out utilizing FTP server (File Transfer Protocol) through Internet3. The FTP server permits sending every kind of files through the web (Internet of other web that utilizes TCP/IP protocol) (Fig. 5) (Fig. 6). There are many programs to connect with a FTP server including Pc or Mac. In our case, the participants just had to connect to the Cytometry Service FTP (ftp.lupo.quimica.uniovi.es) accessing to the "incoming" directory and leaving the .PCX files generated by the DNA software. They entered as "anonymous" users and they used as the password their electronic address (E-mail).
4. Comparative analysis
Once the results were received, the reference center proceeded to evaluate them, comparing the means obtained by the t test for paired data. There were no significant differences in the DNA Index and S-phase fraction parameters. It was observed a complete agreement in DNA Index values in eight of ten exchange cases. In the assessment of S phase fraction the variability was found to be less than 5% in those cases that were analyzed by all the participants.
In the future, it is foreseen that not only will the capacities of the new FCS 3.0 format4 be utilized but also improve the aspects of network communication such us creating a environment of statistical treatment for results, establishing forums of discussion, self-examination and inclusion of forward vs. side scatter diagrams to help in interpretation5.
1. Coon JS et al. Interlaboratory variation in DNA flow cytometry. Results of the College of American Pathologists´ survey. Arch Pathol Lab Med 118:681-685, 1994.
2. Haroske G, Meyer W, Theissig F, Kunze KD. Remote quantitation server for quality assurance in DNA ploidy analysis. 11th International Congress on Diagnostic Quantitative Pathology. Siena, Italy, October 2-4, 1997.
3. Hahn H, Stout R. The Internet Complete Reference. Osborne McGraw-Hill, Berkeley, CA, USA, 1994.
4. Samer LC et al. Proposed new data file standard for flow cytometry, version FCS 3.0. Cytometry, 28:118-122, 1997.
5. Ormerod MG, Titley JC, Imrie PR. Use of light scatter when recording a DNA histogram from paraffin-embedded tissue. Cytometry 21(3):294-299, 1995.